vcf_split.py: VCF Split Function¶
As a single VCF may include the variant sites of multiple loci, it is often necessary to seperate the loci from the VCF. Given a VCF file and a file of loci (i.e. BED or PPP-created statistic file), vcf_split will generate a VCF for each locus.
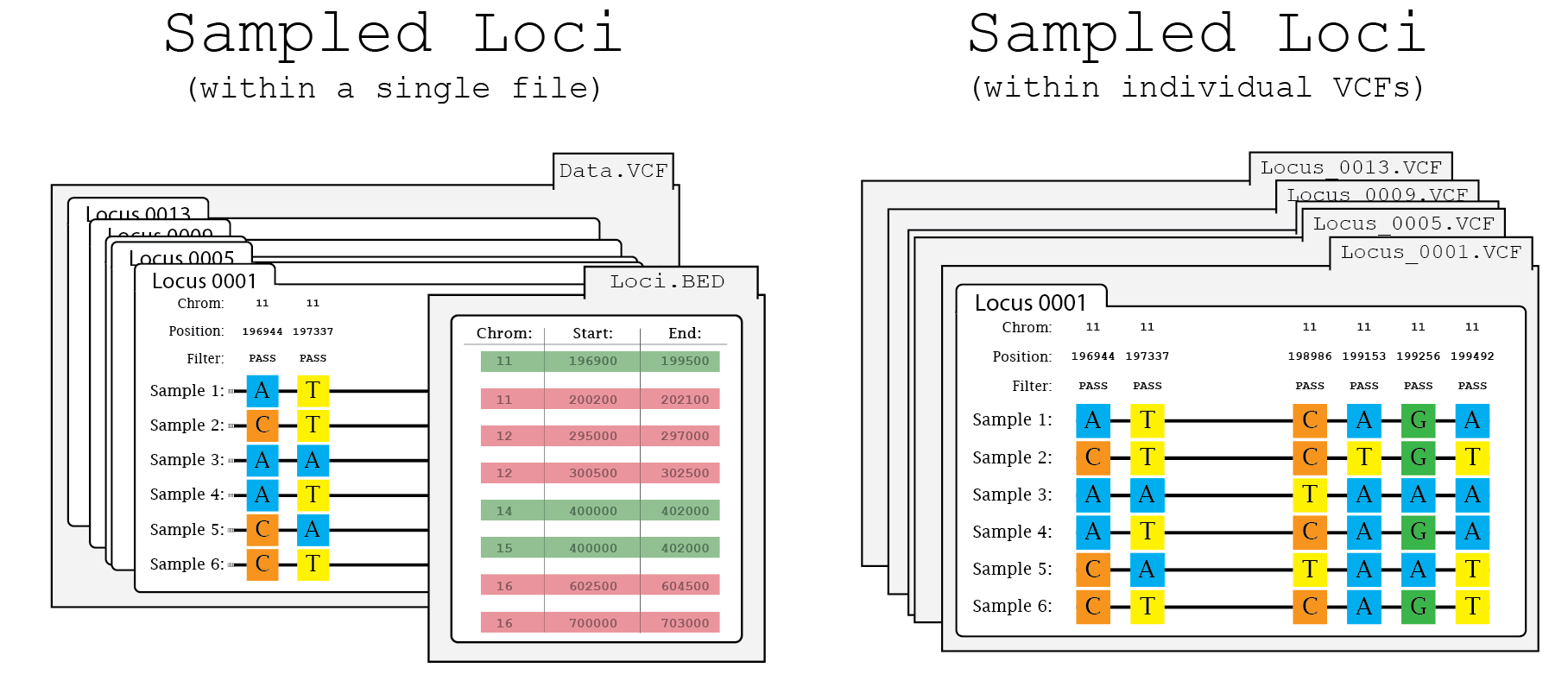
In this illustration of the splitting process, Data.VCF includes variant sites associated with a discrete set of loci (i.e. Locus_0001 - Locus_0013). Once split, a single file (e.g. Locus_0001.VCF) will only contain the variant sites associated with that locus.
Command-line Usage¶
The VCF splitter may be called using the following command:
vcf_split.py
Example usage¶
Command-line to split using a statistic file:
vcf_split.py --vcf examples/files/merged_chr1_10000.vcf.gz --split-file examples/files/sampled.windowed.weir.fst.tsv --split-method statistic-file --model-file examples/files/input.model --model 2Pop
Input Command-line Arguments¶
- --vcf <input_filename>
- Argument used to define the filename of the VCF file to be split.
- --split-file <split_filename>
- Argument used to define the file to be split
- --model-file <model_filename>
- Argument used to define the model file. Please note that this argument cannot be used with the --pop-file argument or individual-based filters.
- --model <model_str>
- Argument used to define the model (i.e. the individual(s) to include and/or the populations for relevant statistics). May be used with any statistic. Please note that this argument cannot be used with --pop-file argument or the individual-based filters.
Output Command-line Arguments¶
- --out-prefix <output_prefix>
- Argument used to define the output prefix (i.e. filename without file extension)
- --out-format <vcf, vcf.gz, bcf>
- Argument used to define the desired output format. Formats include: uncompressed VCF (vcf); compressed VCF (vcf.gz) [default]; and BCF (bcf).
- --out-dir <output_dir_name>
- Argument used to define the output directory.
- --overwrite
- Argument used to define if previous output should be overwritten.
Split Command-line Arguments¶
- --split-method <statistic-file, bed>
- Argument used to define the splitting method. Users may spilit using either a statistic-file (statistic-file) from VCF Calc (or other methods) or a BED file (bed).
- --statistic-window-size <statistic_window_int>
- Argument used to define the size of window calculations. This argument is only required if the BIN_END column is absent within the file.
- --no-window-correction
- Argument used to define if a window should not be corrected to avoid an overlap of a single position (i.e. 100-200/200-300 vs. 100-199/200-299).
Filter Command-line Arguments¶
If using an unfiltered VCF file (e.g. reduce the creation of unnecessary large files) the VCF calculator is able to use either a kept or removed sites/BED file and the individual-based paramemeters.
Individual-Based Arguments¶
Please note that all individual-based arguments are not compatible with either the --model or --model-file command-line arguments.
- --filter-include-indv <indv_str> <indv1_str, indv2_str, etc.>
- Argument used to define the individual(s) to include. This argument may be used multiple times if desired.
- --filter-exclude-indv <indv_str> <indv1_str, indv2_str, etc.>
- Argument used to define the individual(s) to exclude. This argument may be used multiple times if desired.
- --filter-include-indv-file <indv_filename>
- Argument used to define a file of individuals to include.
- --filter-exclude-indv-file <indv_filename>
- Argument used to define a file of individuals to exclude.
Position-Based Arguments¶
- --filter-include-positions <position_filename>
- Argument used to define a file of positions to include within a tsv file (chromosome and position).
- --filter-exclude-positions <position_filename>
- Argument used to define a file of positions to exclude within a tsv file (chromosome and position).
- --filter-include-bed <position_bed_filename>
- Argument used to define a BED file of positions to include.
- --filter-exclude-bed <position_bed_filename>
- Argument used to define a BED file of positions to exclude.