model_creator.py: Model File Creator¶
Many PPP functions were designed to automatically assign relevant populations and/or individuals using a Model file. To enable this functionality, the model_creator.py function may be used to produce Model files by either: i) manually entering the necessary information or ii) by using files with the relevant information. It is also all possible to create multiple models simultaneously and assign populations to more than a single model.
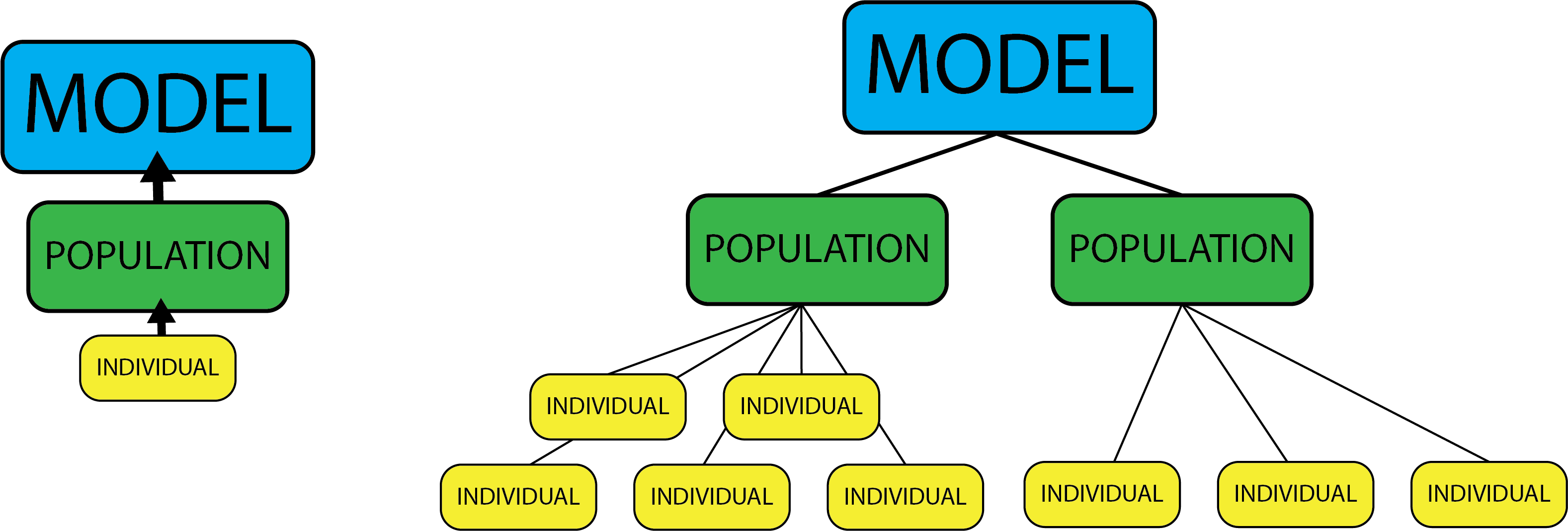
A simple way to visualize models are as a hierarchy. Each Model may contain one or more Populations and each Population may contain one or more Individuals.
Command-line Usage¶
The model creator may be called using the following command:
model_creator.py
Example 1: Simple Model¶
A basic model only require a single population (pop) with a single in individual (ind). Only three commands are required:
- Create and name a model: --model 1Pop
- Assign a pop to a model: --model-pop 1Pop Paniscus
- Assign an ind to a pop: --pop-ind Paniscus Pan_paniscus-9731_LB502
model_creator.py --model 1Pop --model-pop 1Pop Paniscus --pop-ind Paniscus Pan_paniscus-9731_LB502
Example 2: Model Using Files¶
A model may also be created using two file options:
- Assign multiple pops to model: --model-pop-file 2Pop 2Pops.txt
- Assign multiple inds to pop: --pop-ind-file Paniscus Paniscus.txt
model_creator.py --model 2Pop --model-pop-file 2Pop examples/files/2Pops.txt --pop-ind-file Paniscus examples/files/Paniscus.txt --pop-ind-file Troglodytes examples/files/Troglodytes.txt
Paniscus
Troglodytes
Pan_paniscus-9731_LB502
Pan_paniscus-A915_Kosana
Pan_troglodytes_troglodytes-A957_Vaillant
Pan_troglodytes_troglodytes-A958_Doris
Example 3: Update Model in Model File¶
A model may be updated if desired using the following options:
- Assign the model file: --model-file input.model
- Assign the model to update: --update-model 2Pop
- Assign a pop to remove from a model: --model-rm-pop 2Pop Troglodytes
- Assign a new or previously created pop to a model: --model-pop 2Pop Schweinfurthii
- Assign multiple inds to pop (if new): --pop-ind-file Schweinfurthii Schweinfurthii.txt
model_creator.py --model-file examples/files/input.model --update-model 2Pop --model-pop 2Pop Schweinfurthii --model-rm-pop 2Pop Troglodytes
Example 4: Update Population in Model File¶
A population may be updated if desired using the following options:
- Assign the model file: --model-file input.model
- Assign the pop to update: --update-pop Paniscus
- Assign a ind to add to a pop: --pop-ind Paniscus Pan_paniscus-Unknown
- Assign a ind to remove from a pop: --pop-rm-ind Paniscus Pan_paniscus-9731_LB502
model_creator.py --model-file examples/files/input.model --update-pop Paniscus --pop-ind Paniscus Pan_paniscus-Unknown --pop-rm-ind Paniscus Pan_paniscus-9731_LB502
Standard Model Command-line Arguments¶
Except for --model-file all other model-based arguments may be used multiple times.
- --model-file <str>
- Argument used to define the name of a model file.
- --model <model_str>
- Argument used to define the name of a model to create.
- --update-model <str>
- Argument used to define the name of a model to update. Allows for: i) tree update and ii) populations to be added.
- --update-pop <str>
- Argument used to define the name of a population to update. Allows for individuals to be added.
- --model-tree <model_str> <newick_str>
- Argument used to assign a population tree to a model, in Newick format.
- --model-tree-file <model_str> <newick_file>
- Argument used to assign a population tree file to a model, in Newick format.
- --model-pop <model_str> <pop_str>
- Argument used to assign a population to a model.
- --model-pops <model_str> <pop1_str> <pop2_str> ..
- Argument used to assign a multiple populations to a model.
- --model-pop-file <model_str> <pop_file>
- Argument used to assign a multiple populations to a model using a file.
- --pop-ind <pop_str> <ind_str>
- Argument used to assign a individual to a population.
- --pop-inds <pop_str> <ind1_str> <ind2_str> ..
- Argument used to assign a multiple individuals to a population.
- --pop-ind-file <pop_str> <ind_file>
- Argument used to assign a multiple individuals to a population using a file.
Model Update: Compatible Command-line Arguments¶
Please note: --update-model is required to update a model.
- --update-model <str>
- Argument used to define the name of a model to update. Allows for: i) tree update and ii) populations to be added and/or removed.
- --model-file <str>
- Argument used to define the name of a model file.
- --model-tree <model_str> <newick_str>
- Argument may be used to replace a population tree, in Newick format.
- --model-tree-file <model_str> <newick_file>
- Argument may be used to replace a population usng a tree file, in Newick format.
- --model-pop <model_str> <pop_str>
- Argument used to assign a population to the model begin updated.
- --model-pops <model_str> <pop1_str> <pop2_str> ..
- Argument used to assign multiple populations to the model begin updated.
- --model-pop-file <model_str> <pop_file>
- Argument used to assign multiple populations to the model begin updated using a file.
- --model-rm-pop <model_str> <pop_str>
- Argument used to remove a population to the model begin updated.
- --model-rm-pops <model_str> <pop1_str> <pop2_str> ..
- Argument used to remove multiple populations to the model begin updated.
- --model-rm-pop-file <model_str> <pop_file>
- Argument used to remove multiple populations to the model begin updated using a file.
Population Update: Compatible Command-line Arguments¶
Please note: --update-pop is required to update a population.
- --update-pop <str>
- Argument used to define the name of a population to update. Allows for individuals to be added.
- --model-file <str>
- Argument used to define the name of a model file.
- --pop-ind <pop_str> <ind_str>
- Argument used to assign a individual to the population begin updated.
- --pop-inds <pop_str> <ind1_str> <ind2_str> ..
- Argument used to assign multiple individuals to the population begin updated.
- --pop-ind-file <pop_str> <ind_file>
- Argument used to assign multiple individuals to thr population begin updated using a file.
- --pop-rm-ind <pop_str> <ind_str>
- Argument used to remove a individual to the population begin updated.
- --pop-rm-inds <pop_str> <ind1_str> <ind2_str> ..
- Argument used to remove multiple individuals to the population begin updated.
- --pop-rm-ind-file <pop_str> <ind_file>
- Argument used to remove multiple individuals to thr population begin updated using a file.
Output Command-line Arguments¶
- --out <output_filename>
- Argument used to define the complete output filename.
- --overwrite
- Argument used to define if previous output should be overwritten.